Aon, Cortassa, 2002
Model Status
This CellML model runs in both OpenCell and COR. The units have been checked and they are consistent. The CellML model "may" replicate the published results - there are no simple results figures plotting substrate concentration against time in the paper. Neither are there any simple figures showing the results of the "standard" model (certain variables are altered in order to generate the figures as described in the figure captions). The model appears to be very sensitive to the initial conditions (concentrations) of the substrates in the pathway. Where these values have not been specified in the paper they have been set to 0.01.
Model Structure
ABSTRACT: In order to investigate the influence of cytoskeletal organization and dynamics on cellular biochemistry, a mathematical model was formulated based on our own experimental evidence. The model couples microtubular protein (MTP) dynamics to the glycolytic pathway and its branches: the Krebs cycle, ethanolic fermentation, and the pentose phosphate (PP) pathway. Results show that the flux through glycolysis coherently and coordinately increases or decreases with increased or decreased levels of polymerized MTP, respectively. The rates of individual enzymatic steps and metabolite concentrations change with the polymeric status of MTP throughout the metabolic network. Negative control is exerted by the PP pathway on the glycolytic flux, and the extent of inhibition depends inversely on the polymerization state of MTP, i.e. a high degree of polymerization relieves the negative control. The stability of the model's steady state dynamics for a wide range of variation of metabolic parameters increased with the degree of polymerized MTP. The findings indicate that the organization of the cytoskeleton bestows coherence and robustness to the coordination of cellular metabolism.
The original paper reference is cited below:
Coherent and robust modulation of a metabolic network by cytoskeletal organization and dynamics, Miguel A. Aon and Sonia Cortassa, 2002, Biophysical Chemistry, 97, 213-231. PubMed ID: 12050011
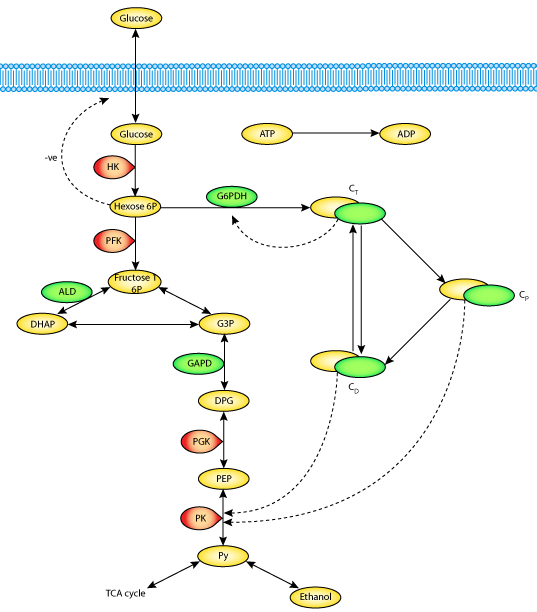 |
| A schematic diagram of the metabolic network, the cycle of assembly-disassembly of microtubular proteins, and their interactions, upon which the Aon-Cortassa model is based. |
