Lovell, Cloherty, Celler, Dokos, 2004
Model Status
This model is currently non-functional.
Model Structure
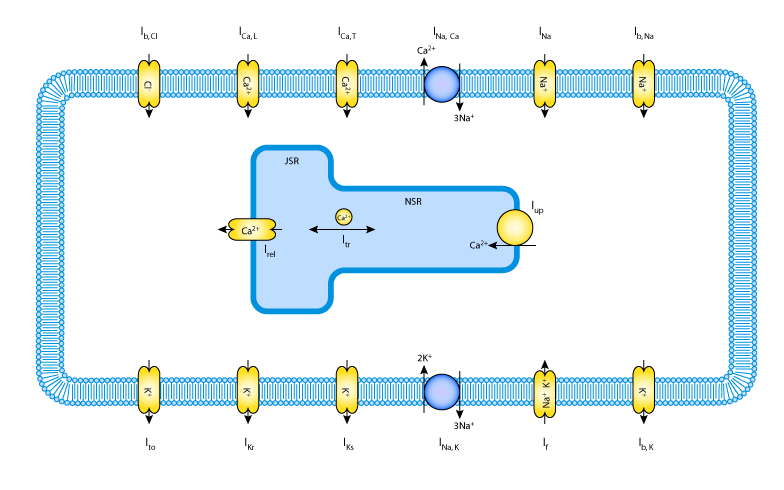
Electrical activity of the healthy mammalian heart is initiated by an area of specialised pacemaker cells in the wall of the heart, known as the sinoatrial node (SAN). These cells are spontaneously active, demonstrating a slow depolarisation to threshold (pacemaker potential) rather than a stable resting potential. Despite heterogeneity amongst the cells of the SAN, the cells are able to synchronise their firing rate and this in turn stimulates the contraction of the atrial myocardium. Synchronisation is achieved by electronic coupling between cells via gap junctions.
To date, two potential theories have been put forward to explain the observed spatial heterogeneity in the action potential characteristics within the SAN:
-
1) The gradient model suggests that the heterogeneity arises intrinsically from a spatial gradient in underlying cellular electrophysiological properties; and
-
2) The mosaic model, which suggests that atrial myocytes are peppered throughout the SAN region, increasing in density towards the periphery. Action potential heterogeneity then arises due to the electronic coupling between atrial and pacemaker myocytes, and the differences between the relative density of the two cell types with the SAN.
In order to further investigate the source of the action potential heterogeneity in the SAN, a mathematical model has been devloped by Lovell et al. (see the figure below). Several mathematical models of the SAN activity have been previously published, including:
-
Demir et al. Sinoatrial Node Model, 1994;
-
Modelling the Ion Currents Underlying Sinoatrial Node Pacemaker Activity, Dokos et al., 1996;
-
A Model of Sinoatrial Node Vagal Control, Dokos et al., 1996;
-
Demir et al. Sinoatrial Node Model, 1999;
-
Zhang et al. Sinoatrial Node Model, 2000;
-
Boyett et al. Sinoatrial Node Model, 2001;
and
-
Kurata et al. Improved Mathematical Model for the Primary Pacemaker Cell, 2002.
Although these models were able to recreate the generic action potential waveforms from the SAN, none were able to represent action potential waveforms from any specific SAN cell. This limits the application of such models to analyse the factors underlying action potential heterogeneity in coupled systems of pacemaker cells, as well as elucidating specific ionic mechanisms underlying action potential heterogeneity in the SAN.
To meet these needs, Lovell et al. have devloped a generic single cell ionic model of rabbit SAN cells, which is based on Markovian state kinetics (all previously published SAN models are based on a Hodgkin-Huxley mechanism). Parameters are fitted to this model using a custom least squares optimisation approach. They then propose a gradient model of the SAN by employing the same optimisation approach to fit a smooth transition in ionic model parameters to achieve a desired variation in AP characteristics.
The model has been described here in CellML. It should be noted that the initial parameters entered into this model are those for the central SAN value. Peripheral SAN values and the interpolation parameters are also provided in the original publication.
The complete original paper reference is cited below:
Progress in Biophysics and Molecular Biology , 85, 301-323. (Full text (HTML) and PDF versions of the article are available to subscribers on the Progress in Biophysics and Molecular Biology website.) PubMed ID: 15142749
 |
