FAIRDO BG example 3.6.cellml
Bond graph example: An enzyme-catalyzed reaction and Michaelis-Menten kinetics
Consider the enzymatic reaction shown below, which is often associated with Michaelis-Menten (MM) kinetics. q1c is a substrate that binds reversibly to an enzyme q3c to form the complex q4c, which breaks down to regenerate the enzyme and yield a product q2c. In conventional MM kinetics this last step is treated as irreversible, since Af2≫Ar2.
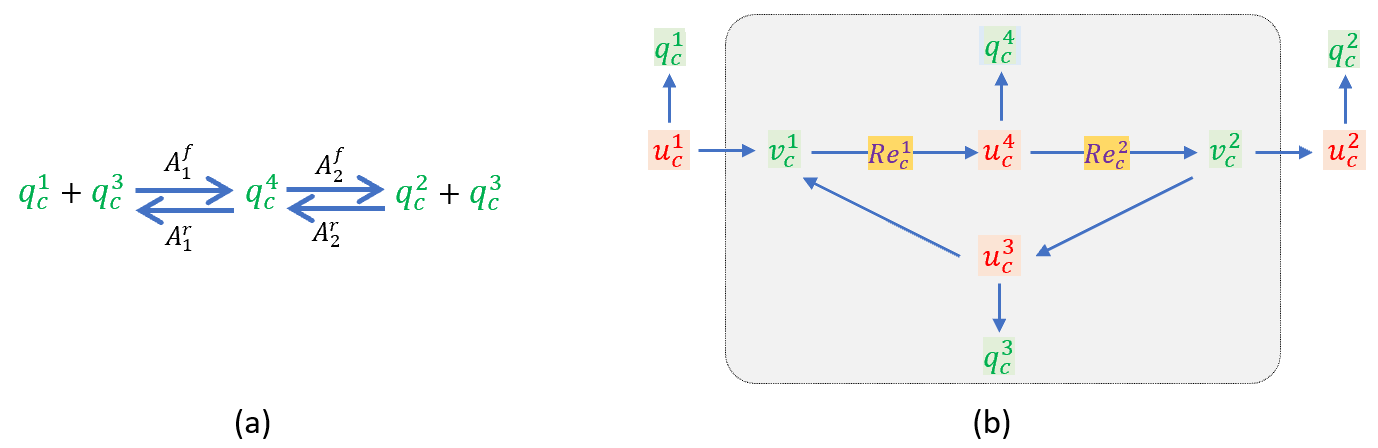
An enzyme (q3c)-catalyzed reaction (a), and its bond graph representation (b). Flux balance is ensured for each of the four species at the 0:nodes, and energy balance at the 1:nodes ensures the correct stoichiometry.
The Views Available menu to the right provides various options to explore this model here in the Physiome Model Repository. Of particular interest is the Launch with OpenCOR menu item, which will load the simulation experiment shown below directly into the OpenCOR desktop application.
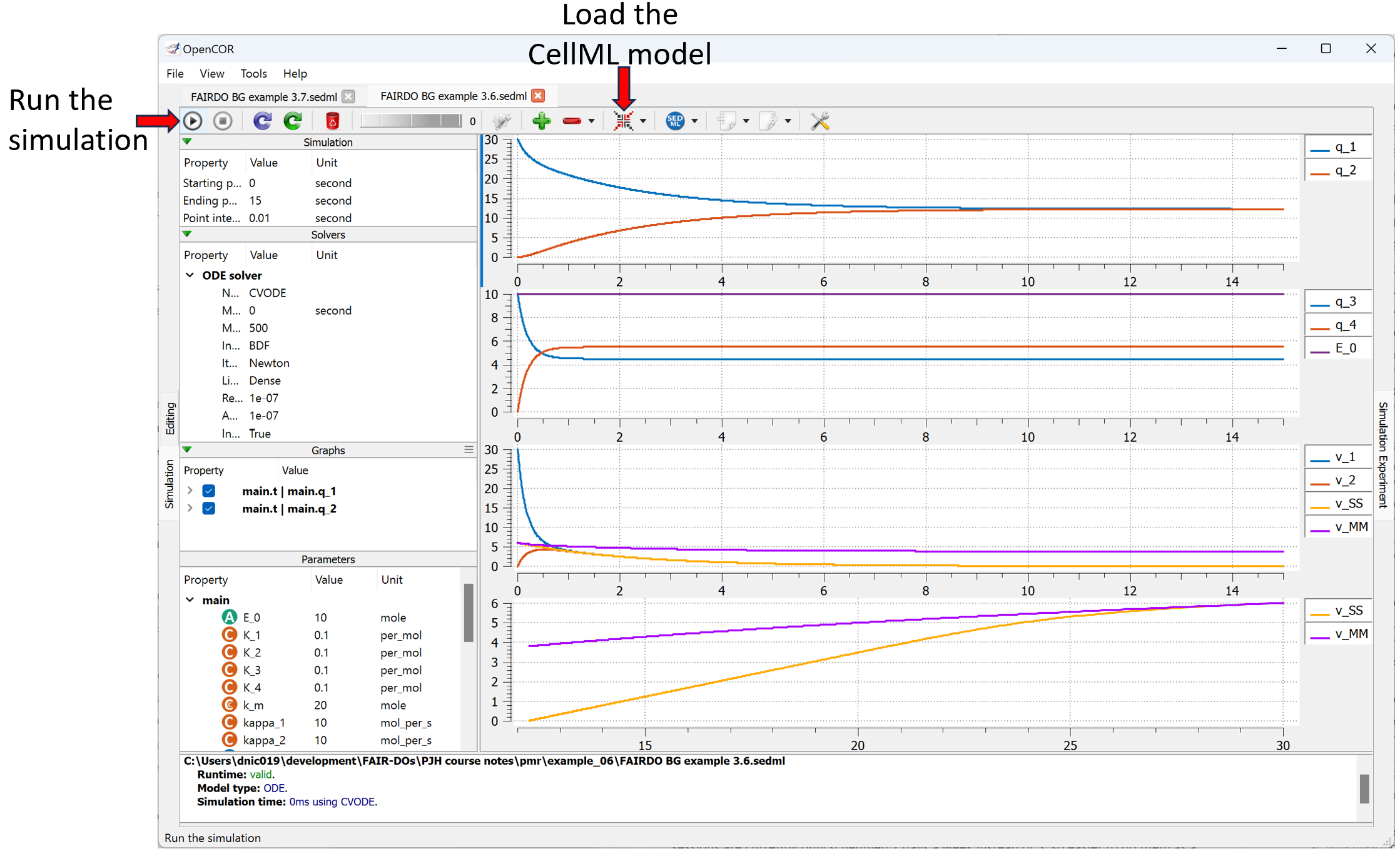
Showing the result of launching the simulation experiment from this exposure in OpenCOR and executing the simulation.
